Context.-Glucose-6-phosphate dehydrogenase (G6PD) deficiency is one of the most common human enzyme deficiencies. More than 130 different molecular abnormalities have been described worldwide, with considerable variation in the enzyme among various racial groups. Data from Kuwaiti populations are scarce, and the studies available are the result of screening male blood donors who may not be truly representative of the Kuwaiti population.
Objective.-The objective of this study was to investigate the mutation spectrum of the G6PD gene among Kuwaiti Arabs.
Design.-DNA was extracted from 82 G6PD-deficient Kuwaiti subjects (75 men and 7 women) and screened for gene mutations using polymerase chain reaction/restriction fragment length polymorphism and polymerase chain reaction/single-strand conformation polymorphism followed by direct sequencing. A total of 1209 randomly selected Kuwaiti adult subjects of both sexes were then screened for any characterized mutation.
Results.-G6PD Mediterranean^sup 563C[arrow right]T^, and A-^sup 202G[arrow right]A,376A[arrow right]G^ genotypes were characterized as the most common variants among the G6PD-deficient population, representing 0.742 and 0.124 allele frequencies, respectively. The 2 previously described mutations, G6PD Chatham^sup 1003G[arrow right]A^ and Aures^sup 143T[arrow right]C^, were found at lower frequencies (0.101 and 0.034, respectively). The allele frequencies for these 4 G6PD variants among the randomly selected Kuwaitis were 0.035, 0.0074, 0.0046, and 0.0023 for Mediterranean, A-, Chatham, and Aures, respectively.
Conclusion.-This study has characterized the molecular heterogeneity of G6PD variants among ethnic Kuwaitis. The findings suggest that gene flow from the Indian sub-continent, sub-Saharan African, and other parts of the Mediterranean may have contributed to the observed G6PD mutations seen in the Kuwaiti population.
(Arch Pathol Lab Med. 2005;129:1144-1147)
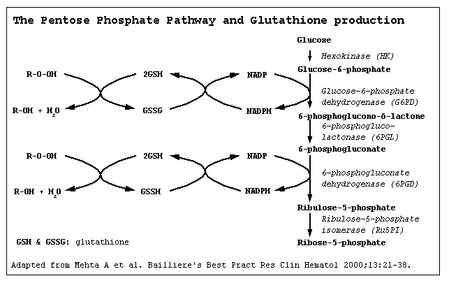
Glucose-6-phosphate dehydrogenase (G6PD) deficiency is one of the most common human enzyme deficiencies, and it is estimated to affect more than 400 million people worldwide.1 Although most of the enzyme-deficient subjects are asymptomatic, deficient individuals may show episodic hemolytic anemia induced by infections or certain drugs and a spontaneous chronic nonspherocytic hemolytic anemia.2 Synthesis of red blood cell G6PD is determined by a gene on the X chromosome, Xq28.3 Dis eases involving this enzyme, therefore, occur more frequently in males than in females. To date, more than 130 different molecular abnormalities and 400 biochemical variants have been described in G6PD-deficient subjects, with a considerable variation in the defect among various racial groups.4 About 13% of the male African American population has a mutant enzyme (G6PD A-) that results in a deficiency of red blood cell G6PD activity to 5% to 15% or less of normal.5 A high incidence (ranging from 5% to 40%) of a variant designated G6PD B- or G6PD Mediterranean is found in Italians, Greeks, and other Mediterranean populations, and in Middle Eastern, African, and Asian ethnic groups.6-8 Several G6PD-deficient variants have been reported among Arabs of the Arabian Gulf region. The most common of these variants is G6PD Mediterranean.9-13 In this study, we have characterized the molecular basis of G6PD deficiency in a large sample of ethnic Kuwaiti subjects. Individuals representative of different districts in Kuwait were further screened to determine the frequency of the mutations in these populations.
SUBJECTS AND METHODS
Sample Collection
Blood samples were collected from 82 known G6PD-deficient subjects (75 men, 7 women) and 1209 randomly selected individuals (660 men, 549 women). Individuals from the 5 different provinces of Kuwait (AlAsema, AlFarwania, AlAhmadi, Allahra, and Hawaii) were included this study. All individuals belong to known Kuwaiti tribes with minimal recent non-Kuwaiti admixture. Consent agreement was provided by all volunteer subjects in this study. The blood samples were taken in EDTA tubes, transported at 4°C, and stored at that temperature for no more than 3 days. Genomic DNA was extracted from peripheral blood leukocytes as previously described.14 Using DNA from the 82 G6PD-deficient patients, the coding region of the G6PD gene encompassing Mediterranean (563 C[arrow right]T) and A- (202 G[arrow right]A, 376 A[arrow right]G) mutations were amplified with polymerase chain reaction (PCR) using primer sets to amplify exons 6, 4, and 5 (Table 1). Polymerase chain reaction assays were performed in 50 µL PCR reaction mix containing 1× Gene Amp PCR buffer, 200 µM each of deoxynucleotide triphosphate, 25 pmol of each primer, 0.5 µg genomic DNA, and 1.25 U of AmpliTaq DNA polymerase. Tubes were heated to 95°C for 5 minutes, then 32 PCR cycles started as follows: 94°C for 1 minute, 56°C or 57°C for 1 minute (Table 1), and 72°C for 1 minute, followed by a final extension step of 72°C for 5 minutes. Ten micro-liters of the amplified fragments were digested overnight at 37°C using the appropriate endonuclease (New England Biolabs, Beverly, Mass). The digestion products were tested on a 3% agarose gel containing ethidium bromide. Digestion patterns of normal and mutant samples are summarized in Table 2.
Samples found to be negative on PCR/endonuclease cleavage analysis were further screened by single-strand conformation polymorphism (SSCP) analysis. The entire coding sequence of the G6PD gene was amplified using specific primers (Table 3). The amplicons were then screened for mutation using nonradioactive SSCP analysis on 0.5 × MDE gels with 10% glycerol in 0.6 × Trisborate-EDTA (TBE) buffer. Gels were run for 16 hours at 4 W and then silver stained. When an electrophoresis mobility shift was observed, the respective PCR products were directly sequenced in the forward and reverse direction on an ABI 3100 automated sequencer, according to the manufacturer's protocol.
DNA extracted from 1209 randomly selected subjects was screened for the presence of C6PD mutations detected in the aforementioned 82 G6PD-deficient individuals using both PCR/ endonuclease analysis and SSCP/direct sequencing.
RESULTS
Of the 82 G6PD-deficient subjects (a total of 89 alleles), 75 (91.5%) were men and 7 (8.5%) were women. G6PD Mediterranean was present in 61 of 82 cases. Of these, we found 54 hemizygotes, 5 homozygotes, and 2 double heterozygotes. This gave a 0.742 allele frequency for this mutation. The G6PD A- mutation showed a frequency of 0.124, with 9 hemizygotes and 2 double heterozygotes who carried G6PD Mediterranean on their second allele. The rest of the G6PD-deficient subjects were all men. They were found to be negative for both G6PD Mediterranean and A-, and were further studied by PCR-SSCP analysis. Band shifts were detected on exon 9 in 9 individuals and on exon 3 of the remaining 3 subjects (Figure). Direct sequencing of the corresponding exons revealed a 1003 G[arrow right]A mutation (G6PD Chatham) on exon 9 and a 143 T[arrow right]C mutation (G6PD Aures) on exon 3. The Aures mutation abolished the recognition site for BglII endonuclease, and this pattern was used to screen for this mutation in the 1209 randomly selected men in the next step. Table 3 summarizes these results.
We screened 1209 subjects (660 men and 549 women, for a total of 1758 chromosomes) for the G6PD Mediterranean, A-, Aures, and Chatham mutations. Polymerase chain reaction/endonuclease cleavage was used to screen for G6PD Mediterranean, A-, and Aures. Mutations in subjects with positive screening results were confirmed by direct sequencing of PCR products. All subjects who were negative for G6PD Mediterranean, A-, and Aures were screened for G6PD Chatham by direct sequencing of exon 9. This step was necessary because SSCP for Chathampositive samples showed a minimal mobility shift that could not be clearly distinguished from normal (Figure). In these cases, direct sequencing of G6PD exon 9 was performed. Table 4 shows the number, frequency, and zygosity of the 4 G6PD mutations identified in the Kuwaiti population. Of the 80 subjects, 45, 28, 5, and 2 were hemizygotes, heterozygotes, homozygotes, and double heterozygotes, respectively. The G6PD mutation frequencies in the Kuwaiti population were 0.068 among men and 0.064 among women, with an overall frequency of 0.066. The G6PD Mediterranean allele was the most common allele, followed by G6PD A-, G6PD Chatham, and G6PD Aures with frequencies of 0.035, 0.0074, 0.0046, and 0.0023, respectively.
COMMENT
G6PD Mediterranean and G6PD A- are the most commonly detected variants among individuals with G6PD deficiency in the Middle Eastern Gulf area, including Iran.6,9-13-15 Screening for the spectrum of G6PD mutations in ethnic Kuwaitis revealed that G6PD Mediterranean (563 C[arrow right]T) is the most common mutation (74.2%), followed by G6PD A- (202 G[arrow right]A; 12.4%). These results are consistent with findings among other Arabic populations in the region.6,9,12,13 These results are slightly different from those previously reported in a Kuwaiti population (G6PD Mediterranean in 72.9% and G6PD A- in 14.3%).11 The results of screening 1209 randomly selected individuals showed that the G6PD A- allele, an allele that predominates in some African populations (including African Americans), occurred with a significantly lower frequency (0.0074) when compared with G6PD Mediterranean (0.035). Whether this indicates some admixture with the continental African population is yet to be determined.
The present study also demonstrated the presence of G6PD Chatham (1003 G[arrow right]A) and G6PD Aures (143 T[arrow right]C) in the Kuwaiti population. G6PD Chatham is now recognized as one of the common variants worldwide.13,15-18 This variant was first identified in a patient of Indian origin, but has also been reported in Algerian Arabs.16,19 Samilchuk et al11 described this mutation in ethnic Kuwaitis, but at a higher frequency. This study included only male blood donors, whereas our study included a large population of male and female subjects, which may give a better representation of the gene frequency in the overall population. The relatively low frequency of this mutation among Kuwaiti subjects may suggest that the mutation is fairly recent in the Kuwaiti population, possibly through links with Oman or the Indian subcontinent. G6PD Aures has been found previously in Kuwait11 and in neighboring populations, including Saudi Arabia3 and the United Arab Emirates.12 This mutation has also been found in the native population of Algeria16,20 and Spain.21 The relative frequency of this mutation in the various populations is low, and the origin of the mutation remains uncertain.
Population-based screening for the common G6PD variants in Kuwait showed an overall frequency of 6.8% among Kuwaiti male chromosomes, which is higher than a previous report that was based on a smaller number of male subjects.10 Nafa et al16 demonstrated that differences in frequency as well as prevalence rates of G6PD deficiency may arise, depending on whether biochemical screening or molecular testing is performed. This might explain the relatively lower prevalence rate of G6PD deficiency reported by Usanga and Ameen22 among Kuwaiti blood donors. An alternative explanation may be that as blood donors were used in the aforementioned study and were from a single area of Kuwait, it is likely that they are not representative of the Kuwaiti population. Furthermore, the Kuwait Central Blood Bank follows the American Association of Blood Banks' strict donor criteria, according to which patients with anemia are deferred for donation; thus, patients with known G6PD deficiency may not be included in a sample of elective blood donors. In addition, G6PD Chatham, the third most common variant in Kuwait, produces a mild or asymptomatic form of G6PD deficiency. Individuals with this variant may have been overlooked during biochemical screening.
We have shown that the overall frequency of G6PD-mutated alleles among the Kuwaiti population is 0.049. The present study included both male and female subjects, whereas that of Samilchuk et al11 revealed G6PD deficiency among 6.5% of only male Kuwaiti blood donors. Kuwait is not a malaria-endemic area, and as G6PD deficiency is known to be associated with malaria,23 this poses some interesting questions as to the origin of these mutations. The results would suggest that there has been significant gene flow from the Indian subcontinent, sub-Saharan African, and other parts of the Mediterranean to Kuwait. Coupled with the relatively short history of Kuwait, this might explain the findings of this study. However, probably the most pertinent part of this study is the clinical significance of the G6PD deficiency, and in particular favism in the population. It has been reported16 that favism is not only associated with G6PD Mediterranean, but may also occur in individuals with G6PD A-. Our study in characterizing the G6PD variant among Kuwaitis is a first step in the rational management of patients with this abnormality.
This work was supported in part by grant NM01/00 from Kuwait University, Sulaibekhat.
References
1. Luzzatto L, Mehta A, Scriver CR, Beaudet AL, Sly WS, eds. Glucose-6-phosphate dehydrogenase deficiency. In: The Metabolic and Molecular Bases of Inherited Disease. New York, NY: McGraw-Hill; 1995:3367-3398.
2. Beutler E. G6PD deficiency. Blood. 1994;84:3613-3636.
3. Beutler E. The molecular biology of G6PD variants and other red cell enzyme defects. Ann Rev Med. 1992;43:47-59.
4. Euzzatto L, Mehta A, Vulliamy TJ. Glucose 6-phosphate dehydrogenase deficiency. In: Scriver CR, Beaudet AL, Sly WS, VaIIe D, eds. The Metabolic and Molecular Bases of Inherited Disease. New York, NY: McGraw-Hill; 2001:4517-4553.
5. Kay AC, Kuhl W, Prchal J, Beutler E. The origin of glucose-6-phosphatedehydrogenase (G6PD) polymorphisms in African-Americans. Am J Hum Genet. 1992;50:394-398.
6. Al-Ali AK, Al-Mustafa ZH, Al-Madan M, Qaw F, Al-Ateeq S. Molecular characterization of glucose-6-phosphate dehydrogenase deficiency in the Eastern Province of Saudi Arabia. Clin Chem Lab Med. 2002;40:814-816.
7. Calabro V, Giacobbe A, Vallone D, et al. Genetic heterogeneity at the glucose-6-phosphate dehydrogenase locus in southern Italy: a study on a population from the Matera district. Hum Genet. 1990:86:49-53.
8. Smith MB. The incidence of glucose-6-phosphate dehydrogenase deficiency in a population of Greek, Italian and Yugoslav origin in Australia. Med J Aust. 1976;25:485-486.
9. El-Hazmi MA, Warsy AS. Glucose-6-phosphate dehydrogenase polymorphism in the Saudi population. Hum Hered. 1986;36:24-30.
10. Samilchuk E, D'Souza B, Al-Awadi S. Population study of common glucose-6-phosphate dehydrogenase mutations in Kuwait. Hum Hered. 1999;49:41-44.
11. Samilchuk E, Al-Suliman I, Usanga E, Al-Awadi S. Clucose-6-phosphate dehydrogenase (G6PD) mutations and UDP-glucuronosyltransferase promoter polymorphism among G6PD deficient Kuwaitis. Blood Cells Mol Dis. 2003;31: 201-205.
12. Bayoumi RA, Nur-E-Kamal MS, Tadayyon M, et a[. Molecular characterization of erythrocyte glucose-6-phosphate dehydrogenase deficiency in Al-Ain district, United Arab Emirates. Hum Hered. 1996;46:136-141.
13. Daar S, Vulliamy TJ, Kaeda J, Mason PJ, Luzzatto L. Molecular characterization of C6PD deficiency in Oman. Hum Hered. 1996;46:172-176.
14. Sambrook J, Fritsch EF, Maniatis T, Nolan C, Ferguson M, eds. Molecular Cloning: Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989:9.16-9.19.
15. Karimi M, Martinez di Montemuros F, Danielli MG, et al. Molecular characterization of glucose-6-phosphate dehydrogenase deficiency in the Pars Province of Iran. Haematologica. 2003:88:346-347.
16. Nafa K, Reghis A, Osmani N, et al. At least five polymorphic mutants account for the prevalence of glucose-6-phosphate dehydrogenase deficiency in Algeria. Hum Genet. 1994;94:513-517.
17. Martinez di Montemuros F, Dotti C, Tavazzi D, Fiorelli G, Cappellini MD. Molecular heterogeneity of glucose-6-phosphate dehydrogenase (G6PD) variants in Italy. Haematologica. 1997;82:440-446.
18. Iwai K, HironoA, Matsuoka H, et al. Distribution of glucose-6-phosphate dehydrogenase mutations in Southeast Asia. Hum Genet. 2001;108:445-449.
19. Vulliamy TJ, D'Urso M, Battistuzzi G, et al. Diverse point mutations in the human glucose-6-phosphate dehydrogenase gene cause enzyme deficiency and mild or severe hemolytic anemia. Proc Natl Acad Sci USA. 1988;85:5171-5175.
20. Nafa K, Reghis A, Osmani N, et al. A new mutation (48 Ile[arrow right]Thr) causing mild G6PD deficiency is associated with favism. Hum Mol Genet. 1993;2:81-82.
21. Rovira A, Vulliamy T, Pujades MA, Luzzatto L, Corrons JL. Molecular genetics of glucose-6-phosphate dehydrogenase (G6PD) deficiency in Spain: identification of two new point mutations in the G6PD gene. Br J Haematol. 1995; 91:66-71.
22. Usanga EA, Ameen R. Glucose-6-phosphate dehydrogenase deficiency in Kuwait, Syria, Egypt, Iran, Jordan and Lebanon. Hum Hered. 2000;50:158-161.
23. Verrelli BC, McDonald JH, Argyropoulos G, et al. Evidence for balancing selection from nucleotide sequence analyses of human G6PD. Am J Hum Genet. 2002;71:1112-1128.
Suad AlFadhli, PhD; Salim Kaaba, PhD; Alaa Elshafey, MD, PhD; Matra Salim, BSc; Anwar AlAwadi, PhD; Layla Bastaki, MD
Accepted for publication May 12, 2005.
From the Department of Medical Laboratory Sciences, Kuwait University, Sulaibekhat (Drs AIFadhli, Kaaba, and AlAwadi, and Ms Salim); and Kuwait Medical Genetic Center, Sulaibekhat (Drs Elshafey and Bastaki).
The authors have no relevant financial interest in the products or companies described in this article.
Corresponding author: Suad AIFadhli, PhD, Kuwait University, Allied Health Sciences, Department of Medical Laboratory Sciences, PO Box 31470, Sulaibekhat-90805, Kuwait (e-mail: s.alfadhli@hsc.edu.kw; suadq8@yahoo.com).
Copyright College of American Pathologists Sep 2005
Provided by ProQuest Information and Learning Company. All rights Reserved