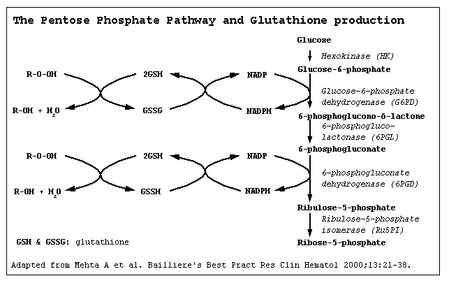
Glucose-6-phosphate dehydrogenase (G6PD) catalyzes the committed step of the pentose phosphate pathway, the only method by which red blood cells can regenerate NADPH (Carson et al. 1956). Over 400 biochemical variants of G6PD deficiency have been described worldwide (Beutler 1990). However, the molecular basis of many of these variants has not been substantiated, and it now appears that the degree of G6PD heterogeneity at the molecular level may not be as high as has been predicted from biochemical analyses (Beutler 1989, 1991, 1992, 1993; Beutler et al. 1991; Vulliamy et al. 1993).
G6PD deficiency is widespread in Papua New Guinea (PNG). Its frequency in males ranges from 0% in many highland groups to more than 50% in the Butibum village in Morobe Province (Yenchitsomanus et al. 1986). Biochemical studies of G6PD deficiency in PNG have revealed a high degree of heterogeneity, with 24 unique variants described to date (Chockkalingam and Board 1980; Chockkalingam et al. 1982; Kirkman et al. 1968; Yoshida et al. 1973). We have proposed previously that many variant forms of G6PD deficiency arose independently in PNG and remained localized in distribution because of the fragmentation of the population (Chockkalingam et al. 1982).
In this study we have investigated the molecular basis of G6PD deficiency in PNG to assess whether the degree of heterogeneity at the molecular level is commensurate with that seen at the biochemical level.
Materials and Methods
Sample Collection. Two different sets of samples were used in this study. Five hundred twelve blood samples, collected from the Maprik (Wosera), Kupiano (Wanigela and Kalo), and National Capital (Koki) districts of PNG, were screened for G6PD deficiency using starch gel electrophoresis (Harris and Hopkinson 1978). Only two males, one from the Wosera and one from the Kalo, were conclusively identified as being hemizygous for G6PD deficiency. In addition, 32 G6PD-deficient samples collected over the years from various parts of PNG were included in this study. Details of the screening of these samples are provided by Chockkalingam et al. (1982) and Yenchitsomanus et al. (1986).
Biochemical Characterization of the Wosera G6PD-Deficient Sample. Additional blood samples were obtained from the Wosera G6PD -- proband and his family members for G6PD enzyme characterization. Blood samples were transported to Canberra, Australia, on ice, and G6PD was purified within 48 hours of collection, as described by Beutler et al. (1968). Biochemical characterization was carried out in accordance with WHO standard protocols (World Health Organization 1967). The Kalo variant was not biochemically characterized because a fresh blood sample could not be obtained.
DNA Analysis of G6PD Coding Exons. The coding exons of the G6PD genes of the two hemizygous deficient (G6PD -) males were sequenced after polymerase chain reaction (PCR) amplification. In addition, the coding exons of a G6PD+ male from the Wosera were also sequenced for comparison. The sequencing strategies resulted in the sequencing of introns 3, 6, 10, 11, and 12.
DNA was prepared from buffy coats using a standard salting-out procedure (Miller et al. 1988). PCR amplification of the coding exons was carried out according to the procedure of Poggi et al. (1990) with some modifications. Briefly, each amplification reaction consisted of 200 ng of genomic DNA, 725 pmol of each primer pair, 10% Taq polymerase buffer (Promega), 10 pmol of each dNTP (Sigma), +/- 10% dimethyl sulfoxide (DMSO), and 2 units of Taq polymerase (Promega) in a 50 mu 1 volume. PCR cycles were modified for each primer pair, as summarized in Table 1.(Table 1 omitted) The sizes of the resultant PCR products were verified by electrophoresis in 2% agarose gels alongside a molecular weight marker (band size range 9-622 bp, MspI digested pBR322 plasmid).
The PCR products were blunt-end ligated into a SmaI cut M13 mp18 vector by standard molecular techniques (Sambrook et al. 1989). Fragments that would not ligate into M13 were ligated into a pGEM-T vector (Promega) using conditions indicated by the manufacturer. The cloned fragment was then excised using flanking restriction enzyme sites and subcloned into complimentary sites in M13 mp18 or mp19. Dideoxy-sequencing (Sanger et al. 1977) was carried out using M13 universal primers and a multiwell microtiter plate DNA sequencing system
T sub 7 DNA polymerase kit (Amersham)
Any nucleotide changes were verified using restriction enzyme digestion of the PCR product or by resequencing of new PCR products where no restriction sites were altered.
Sequence-Specific Oligonucleotide Hybridization, To assess the distribution of the Wosera and Kalo variants in PNG, we screened the DNA from 32 Papua New Guineans with G6PD deficiency from across the country (Figure 1) using sequence-specific oligonucleotide (SSO) hybridization.(Figure 1 omitted) Genomic DNA in the region of the observed mutations was amplified using the G6P11F and G6P11B primer pair, which resulted in a 665-bp PCR product (Figure 2).(Figure 2 omitted) PCR products were electrophoretically separated on 0.8% agarose gels and transferred onto nylon membrane (Hybond N+, Amersham) by sandwich blotting (Gao 1992). Hybridization and washing of SSO filters were carried out according to the procedures described by Gao (1992). The membranes were probed with four chemically synthesized 19mer oligonucleotides (SSO probes) designed to span the two nucleotide changes and their G6PD + B variant counterparts (Table 2).(Table 2 omitted) The SSO probes were end-labeled by gamma sup 32 P-ATP (4000 Ci/mmol; Bresatec) using T4 polynucleotide kinase (Pharmacia). DNA samples from individuals with the Wosera G6PD-, +/-, and + genotypes and the Kalo G6PD -- hemizygote were used as hybridization controls. The presence of an NlaIII polymorphism in intron 11 was also assessed by restriction analysis and SSO hybridization (Figures 3 and 4).(Figures 3 and 4 omitted)
Results
Wosera Variant. A summary of the biochemical properties of the Wosera G6PD -- enzyme is shown in Table 3.(Table 3 omitted) Although other PNG variants share some biochemical properties with the Wosera variant (see Table 3), the variant is unique because it does not utilize NAD and shows no change in electrophoretic mobility.
The Wosera G6PD + coding sequence was identical to the G6PD B sequence (Persico et al. 19X). Therefore variations in the coding sequence of the two G6PD deficient genes were assumed to be responsible for the enzyme deficiencies.
The Wosera G6PD -- variant gene contained a G to A transition at nt1388 within exon 12, resulting in an Arg sup 463 -- Cys substitution. The genetic transmission of the nt1388 sup A mutation was confirmed by hybridization of the nt1338 sup A -specific oligonucleotide (EX12W) to the proband's mother and brother (Figure 4). This mutation has also been described previously as G6PD Kaiping in samples of Asian origin (Chui et al. 1991).
A polymorphic NlaIII site was detected in intron 11, and SSO hybridization indicated that the Wosera G6PD -- samples possessed the NlaIII (T) allele (Figure 3). This particular polymorphism has also been previously reported outside PNG (Vulliamy et al. 1991).
Kalo Variant, The Kalo G6PD -- variant gene contained a C to T transition at position nt1360 within exon 11, resulting in an Arg454 -- Cys substitution. The nt1360 sup T mutation eliminates an HhaI site and was confirmed by direct digestion of the amplified PCR product (Figure 3). This mutation has been described previously as G6PD Union, which apparently has worldwide distribution (Rovira et al. 1994). The Kalo G6PD -- sample was found to have the intron 11 NlaIII + (C) allele.
Thirty-two G6PD -- samples from seven additional PNG populations were screened for the presence of the nt1360 sup T and nt1388 sup A G6PD variants. Two of the samples from Aroana and Kaparoko (Central Province) and one from Madang (Madang Province) were found to carry the nt1360 sup T mutation seen in the Kalo variant (Figure 1; Table 4).(Table 4 omitted) These samples also carried the NlaIII + (C) allele of the intron 11 polymorphism. None of the screened G6PD-deficient samples contained the nt1388 sup A Wosera mutation.
Discussion
We have characterized the G6PD -- genes of two individuals from different geographic regions of PNG (Wosera and Kalo) and have determined that the molecular basis of their deficiency is different. The nt1388 sup A mutation found in the Wosera G6PD (-) gene is the same as the substitution responsible for G6PD Kaiping found commonly in Chinese populations (Chang et al. 1992; Chui et al. 1991, 1993; Tang et al. 1992). In one Chinese study the nt1388 sup A mutation accounted for up to 21% of G6PD-deficient individuals (Chang et al. 1992). Despite the identical nucleotide substitutions, the biochemical characterization of the Wosera G6PD -- variant did not suggest that it was G6PD Kaiping. In fact, a comparison of the Wosera G6PD variant and five Chinese biochemical variants (G6PD Anant, G6PD Kaiping, G6PD Sapporo-like, G6PD Dhon, and G6PD Petrich-like), which all share the single G -- A transition at nt1388 (Chui et al. 1991), revealed considerable biochemical variation. This comparison provides further evidence that G6PD variants with the same molecular basis do not necessarily yield the same biochemical profile. This may be due to the extremely labile nature of the G6PD enzyme, resulting in varying levels of biochemical degradation during storage and transport of samples. Similarly, although the Wosera G6PD variant appears to be biochemically unique, other PNG variants that share some biochemical similarity to it
e.g., G6PD Wewak, G6PD Kar Kar, and GGPD Manus (see Table 3)
may possess the nt1388 sup A substitution as well. Molecular analysis of these previously described G6PD variants is required to confirm whether they do in fact have the same molecular lesion.
The G6PD Union nt1360 sup T mutation found in Kalo has previously been noted in individuals of Italian (Calabro et al. 1993), Spanish (Rovira et al. 1994), Chinese (Perng et al. 1992), Laotian, Filipino (Beutler et al. 1992b), Japanese, African American (Hsia et al. 1993), and Vanuatuan descent (M. Ganczakowski and L. Luzzatto, personal communication, 1994). It is likely that this widespread but sporadic distribution of the G6PD Union mutation is the result of the nt1360/1 CpG dinucleotide being a mutational hot spot (Rovira et al. 1994), resulting in the nt1360 sup T mutation arising independently in populations from different regions of the world.
Screening of G6PD-deficient individuals from seven other PNG populations was carried out to provide an idea of the distribution of the nt1360 sup T and nt1388A mutations in PNG (Figures 3 and 4). The presence of the Kalo variant in three other population samples (Aroana, Kaparoko, and Madang) suggests that the nt1360 sup T mutation is widespread. It appears to be particularly common along the south coast of PNG, with three out of three G6PD -- genes carrying this mutation (Figure 1). The Wosera nt1388 sup A mutation on the other hand was not found elsewhere in PNG and may therefore have a limited distribution. The remaining 29 G6PD -- samples do not contain either of these variants, suggesting the presence of at least one further molecular variant of G6PD deficiency in PNG.
A concurrent study of G6PD deficiency in Melanesians indicated the presence of the nt1360 sup T mutation in Vanuatu. The same study described three new G6PD variants, one of which (nt383 sup C ) was found in a male who had migrated from the north coast of PNG (Ganczakowski 1995). It will be interesting to screen the 29 remaining G6PD -- samples for the presence of the Vanuatuan G6PD variants to determine whether they are also widely distributed.
Because both G6PD deficiencies characterized at the molecular level in PNG are found in Asian populations, it is possible that the G6PD-deficient genes may have been introduced through immigration from Asia to PNG (Serjeantson 1989). If this was a recent event, the G6PD variants found in PNG might be expected to share the same G6PD haplotype as those from Asia. It was noted that the Wosera nt1388 sup A variant is present on the same NlaIII -- (T) haplotype as all the nt1388 sup A variants seen in Taiwan (D.T.Y. Chui, personal communication, 1994). However, the T allele occurs frequently in most populations studied so far (Beutler et al. 1992a), including PNG. Therefore it is still possible that the nt1388A variant may have arisen independently in PNG on a G6PD gene carrying the T allele.
To date, the only haplotype information provided for nt1360 sup T variants is from Spanish Gypsies and Oriental samples that have an intron 11 NlaIII (T) haplotype (Rovira et al. 1994). On the other hand, the Kalo variant and three other PNG nt1360 sup T variants all have an NlaIII + (C) haplotype, suggesting a separate origin. Haplotype analysis of those nt1360 sup T variants found closer to PNG (e.g., in the Philippines and Vanuatu) and molecular characterization of the remaining G6PD -- samples from PNG should provide a clearer picture of the regional influences over G6PD deficiencies in PNG.
Acknowledgments The Wosera and Koki blood samples were collected by George Koki, for which we are extremely grateful. Initial G6PD screening of the Wosera and Kalo populations was carried out by Stephan Pluschke. We would also like to thank the family of the subject with the Wosera G6PD -- variation for agreeing to provide additional blood samples and Taka Suzuki and Sue Serjeantson for their help and advice.
1 Institute of Cell, Animal, and Population Biology, University of Edinburgh, Edinburgh EH9 3JT, Scotland.
2 Aboriginal and Torres Strait Islander Health Unit, Australian Institute of Health and Welfare. GPO Box 570, Canberra, ACT 2601, Australia.
3 Human Genetics Group, John Curtin School of Medical Research. Australian National University, Canberra, ACT 2601, Australia.
4 Molecular Genetics Group, John Curtin School of Medical Research, Australian National University, Canberra, ACT 2601, Australia.
Literature Cited
Beutler, E. 1989. Glucose-6-phosphate dehydrogenase: New perspectives. Blood 73:1397-1401.
Beutler, E. 1990. The genetics of glucose-6-phosphate dehydrogenase deficiency. Semin. Hematol. 27:137-164.
Beutler, E. 1991. Glucose-6-phosphate dehydrogenase deficiency. New Engl. J. Med. 324:169-174.
Beutler, E. 1992. The molecular biology of the G6PD variants and other red cell enzyme defects. Annu. Rev. Med. 43:47-59.
Beutler, E. 1993. Study for glucose-6-phosphate dehydrogenase: History and molecular biology. Am. J. Hematol. 42:53-58.
Beutler, E., C.K. Mathai, and J.E. Smith. 1968. Biochemical variants of glucose-6-phosphate dehydrogenase giving rise to congenital nonspherocytic hemolytic disease. J. Hematol.
Beutler, E., W. Kuhl, T. Gelbart et al. 1991. DNA sequence abnormalities of human glucose-6-phosphate dehydrogenase variants. J. Biol. Chem. 266:4145-4150.
Beutler, E., B. Westwood, and B. Sipe. 1992a. A new polymorphic site in the G6PD gene. Hum. Genet. 89:485486.
Beutler, E., B. Westwood, W. Kuhl et al. 1992b. Glucose-6-phosphate dehydrogenase variants in Hawaii. Hum. Hered. 42:327-329.
Calabro, V., P.J. Mason, S. Filosa et al. 1993. Genetic heterogeneity of glucose-6-phosphate dehydrogenase deficiency revealed by single-strand conformation and sequence analysis. Am. J. Hum. Genet. 52:527-536.
Carson, P.E., F.C. Larkin, C.E. Ickes et al. 1956. Enzymatic deficiency in primaquine-sensitive erythrocytes. Science 124:484--485.
Chang, J.G., S.S. Chiou, L.1. Perng et al. 1992. Molecular characterization of glucose-6-phosphate dehydrogenase (G6PD) deficiency by natural and amplification created restriction sites: Five mutations account for most G6PD deficient cases in Taiwan. Blood 80:1079-1082.
Chockkalingam, K., and P. Board. 1980. Further evidence of heterogeneity of glucose-6-phosphate dehydrogenase deficiency in Papua New Guinea. Hum. Genet. 56:209-212.
Chockkalingam, K., P. Board, and G. Nurse. 1982. Glucose-6-phosphate dehydrogenase deficiency in Papua New Guinea: The description of 13 new variants. Hum. Genet. 60:189-192.
Chui, D.T.Y., L. Zuo, L. Chao et al. 1993. Molecular characterization of glucose-6-phosphate dehydrogenase (G6PD) deficiency in patients of Chinese descent and identification of new base substitutions in the human G6PD gene. Blood 81:2150-2154.
Chui, D.T.Y., L. Zuo, E. Chen et al. 1991. Two commonly occurring nucleotide base substitutions in Chinese G6PD variants. Biochem. Biophys. Res. Commun. 180:988-993.
Ganczakowski, M., M. Town, D.K. Bowden et al. 1995. Multiple glucose-6-phosphate dehydrogenase deficient variants correlate with malaria endemicity in the Vanuatu archipelago (southwestern Pacific). Am. J. Hum. Genet. 56:294-301.
Gao, X. 1992. Nucleotide sequence diversity of HLA class II genes in Australian Aborigines and populations of Asia-Oceania. Ph.D. dissertation, Australian National University, Canberra.
Harris, H., and D.A. Hopkinson. 1978. Handbook of Enzyme Electrophoresis in Human Genetics. Amsterdam, Netherlands: North Holland.
Hsia, Y.E., F. Miyakawa, J. Baltazar et al. 1993. Frequency of glucose-6-phosphate dehydrogenase (G6PD) mutations in Chinese, Filipinos, and Laotians from Hawaii. Hum. Genet. 92:470-476.
Kirkman, H.N., C. Kidson, and M. Kennedy. 1968. Variants of human glucose-6-phosphate dehydrogenase: Studies of samples from New Guinea. In Hereditary Disorders of Erythrocyte Metabolism. E. Beutler, ed. New York Grune and Stratton. 126-145.
Miller, S.A., D.D. Dykes, and H.F. Polesky. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16:1215.
Perng, L.I., S.S. Chiou, T.C. Liu et al. 1992. A novel C to T substitution at nucleotide 1360 of cDNA which abolishes a natural HhaI site accounts for a new G6PD deficiency gene in Chinese. Hum. Molec. Genet. 1:205.
Persico, M.G., G. Viglietto, G. Martini et al. 1986. Isolation of human glucose-6-phosphate dehydrogenase (G6PD) cDNA clones: Primary structure of the protein and unusual 5' noncoding region. Nucleic Acids Res. 14:2511-2522.
Poggi, V., M. Town, N.S. Foulkes et al. 1990. Identification of a single base change in a new human mutant glucose-6-phosphate dehydrogenase gene by polymerase chain reaction amplification of the entire coding region from genomic DNA. Biochem. 271:157-160.
Rovira, A., T.J. Vulliamy, A. Pujades et al. 1994. The glucose-6-phosphate dehydrogenase (G6PD) deficient variant G6PD Unin (454 Arg Cys) has a worldwide distribution possibly due to recurrent mutation. Hunz. Molec. Genet. 3:833-835.
Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Biology: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, v. 1, 2.82-2:107.
Sanger, F., S. Nicklen, and A.R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467.
Serjeantson, S.W. 1989. The colonization of the Pacific: The genetic evidence. In The Colonization of the Pacific: A Genetic Trail, A.V.S. Hill and S.W. Serjeantson, eds. Oxford, England: Clarendon Press, 286-294.
Tang, T.K., C.S. Huang, M.J. Huang et al. 1992. Diverse point mutations result in glucose-6-phosphate dehydrogenase (G6PD) polymorphism in Taiwan. Blood 79:2135-2140.
Vulliamy, T., E. Beutler, and L. Luzzatto. 1993. Variants of glucose-6-phosphate dehydrogenase are due to missense mutations spread throughout the coding region of the gene. Hum. Mutat. 2:159-167.
Vulliamy, T., A. Othman, M. Town et al. 1991. Polymorphic sites in the African population detected by sequence analysis of the glucose-6-phosphate dehydrogenase gene outline the evolution of the variants A and A-. Proc. Natl. Acad. Sci. USA 88:8568-8571.
World Health Organization. 1967. Standardization of Procedures for the Study of Glucose-6-Phosphate Dehydrogenase. Technical Report Series 366. Geneva, Switzerland: World Health Organization.
Yenchitsomanus, P., K.M. Summers, K. Chockkalingam et al. 1986. Characterization of G6PD deficiency and thalassemia in Papua New Guinea. Papua New Guinea Med. J. 29:53-58.
Yoshida, A., and E. Beutler. 1978. Human glucose-6-phosphate dehydrogenase variants: A supplementary tabulation. Ann. Hum. Genet. 41:347-355.
Yoshida, A., E.R. Giblett, and L.A. Malcolm. 1973. Heterogeneous distribution of glucose-6-phosphate dehydrogenase variants with enzyme deficiency in the Markham valley area of New Guinea. Ann. Hum. Genet. 37:145-150.
Copyright Wayne State University Press Jun 1996
Provided by ProQuest Information and Learning Company. All rights Reserved