Typically, one of the first sure signs of a failing public health system is the spread of food-borne infections. In the U.S., however, the emergence and reemergence of food-borne pathogens is due mostly to changes in dietary habits, alterations in food processing and packaging, the globalization of our food supply, microbial mutation, as well as the development of new pathogenic agents.
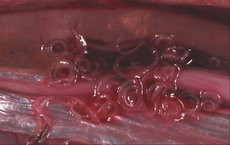
Americans' shift from eating red meat to seafood has produced several outbreaks of hepatitis A virus, Norwalk virus, Aeromonas, and Vibrio species. The increasing popularity of eating raw or partially cooked seafood (customary in Japan) has led to the introduction of human anisakiasis into this country, a food-borne disease caused by the cod worm or herring worm.
The increasing incidence of Salmonella infection in the U.S. certainly can be attributed to the introduction of large-scale poultry rearing and processing facilities. While the introduction in 1967 of plastic wraps for packaging fresh mushrooms helped to preserve this highly perishable food, the high respiratory rate of the product rapidly produced an anaerobic environment ideal for Clostridium botulinum to grow. After a series of botulism outbreaks, two small holes for oxygen transfer were added to the packaging to prevent C botulinum from thriving.
There are various points in the farm-to-consumer chain where food safety is compromised. For instance, the extensive use of antibiotics for livestock to enhance growth and reduce/prevent illness has been a source of great controversy. Many scientists and public health professionals worry this practice could lead to the development of resistant organisms, which would then be passed along to human populations.
This article highlights only a few of the many food-borne pathogens currently requiring laboratorians' immediate attention. These life-threatening organisms include Escherichia coli, Salmonella, Listeria monocytogenes, and Helicobacter pylori.
On January 13, 1993, a physician in Washington State reported a cluster of children with hemolytic uremic syndrome (HUS), the major cause of acute kidney failure in pediatric patients in this country. Also noted was a dramatic increase in emergency room visits for bloody diarrhea in patients of all ages. These events proved to be merely the tip of the iceberg. When this multistate outbreak was over, more than 500 children and adults had become ill; for four children, the illness was fatal.
The culprit was traced back to hamburger patties contaminated with the bacterial pathogen Escherichia coli 0157:H7. This outbreak, eventually traced back to a national fast-food chain, implicated at least 93 of its restaurants. While the specific meat was traced to one processing plant, investigators were unable to determine the meat source.
During the slaughter process, intestinal fluid or feces of infected cattle can drip onto the surface of the meat, contaminating it. It is theorized the harmful bacteria on the surface of the raw meat become mixed throughout the meat during the grinding process. One hamburger patty can contain the meat from as many as 20 cows.
Following this 1993 outbreak, the hamburger was vilified in the media. Public health authorities issued warnings about cooking ground beef, specifically, that the center of hamburger patties should contain no visible pink and the meat should be cooked until the juices run clear. The U.S. Department of Agriculture estimated up to 3% of raw hamburger meat sold in supermarkets could be contaminated.
LIFETIME FRIEND TURNED ENEMY
E coli bacteria are short Gram-negative rods that are part of the normal flora of the intestines of most warm-blooded animals. They colonize the digestive tract a few hours after birth and essentially remain there for life, helping us to digest our food and staying out of trouble unless they somehow are transferred to a body site (e.g., the urinary tract) where they don't belong. These organisms are the most common facultative anaerobe in the large bowel and provide protection against colonization by other harmful microbes. There are, though, five distinct groups of E coli that can cause enteric disease: 1) enteroinvasive, 2) enteropathogenic, 3) enterotoxigenic, 4) enteroadherent, and 5) enterohemorrhagic.
Today E coli 0157:H7 is a subset of the enterohemorrhagic variety that produces Shiga-like toxins (verotoxins) 1 and 2. It has become a prevalent emerging food-borne pathogen in the U.S., more common than Shigella and Yersinia. Common sources of infection include beef, raw milk, untreated water, as well as unpasteurized apple cider and dry-cured salami. Person-to-person transmission is extremely common since the organism has a very low infective dose and can act like Shigella with explosive rapid transmission, particularly in day care centers and geriatric facilities.[1]
The toxin produced by E. coli 0157:H7 in the intestines can cause anything from a mild diarrhea to severe hemorrhagic colitis, where the cells of the intestinal lining are damaged, allowing blood to pass into stool. When a patient's bloody diarrhea is accompanied by stomach pain, nausea, and vomiting, health care professionals should immediately add E coli 0157:H7 to their list of suspects. From 2%-7% of these illnesses progress to HUS, where the toxin enters the bloodstream through the damaged intestinal wall and travels to the smaller arteries that supply the kidneys, going on to damage those vessels. HUS is fatal in about 3%-5% of these cases.[1-3]
Distribution of E coli 0157:H7 probably is worldwide; the majority of cases have been noted in North America and Europe. Many of these infections go unrecognized since many labs don't look for it routinely. The incubation period for this pathogen is rapid (12-60 hours), and oral replacement of fluids and electrolytes is the most important treatment. Most scientists believe the organism emerged as an enteric pathogen by acquiring a gene for Shiga-like toxin production spread by a bacterial virus and infecting an otherwise unremarkable host E. coli. (See Figure 1 for a time line depicting how E. coli. has spread throughout the U.S.)
TESTING FOR E. COLI 0157:H7
E coli 0157:H7 should be reported to public health authorities immediately following its isolation. As with other enteric pathogens, stool specimens should be obtained as soon as possible after the onset of diarrhea.
While it was previously believed most patients secreted the organism for less than one week following the onset of symptoms, recent studies indicate 38% of patients may still be positive three weeks following initial infection. One such study indicated possible isolations from 58% of infected children after three weeks, yet only 8% of adults were still positive after this time interval.
Isolation and presumptive ID. Any lab attempting to isolate E coli 0157:H7 must use the screening medium MacConkey with Sorbitol agar (SMAC), which incorporates sorbitol in place of lactose. Since more than 90% of all E coli strains use sorbitol, and nearly all isolates of E coli 0157:H7 do not, this medium is the method of choice for 0157 screening and isolation.
A minimum of five sorbitol-negative colonies should be agglutinated with 0157 antisera or latex agglutination reagents. Positive colonies may be presumptively reported as E coli 0157. Lactose and lysine decarboxylase also may be used to delineate the organism; the vast majority tested have been found to be positive in both substrates.
Confirming 0157:H7 requires tube titration agglutination of 0157 antisera to the appropriate titer (as indicated by the serum manufacturer) and identification of the H7 flagellar antigen. Unfortunately, H7 identification is not as straightforward as 0157 analysis. Most 0157:H7 isolates require a minimum of at least three to five serial passages through 2% motility medium before sufficient H7 antigen is present for confirmation. A few may be positive after only two passes, and some require more than 10.[1]
Use of MUG. Some labs test E coli 0157:H7 strains for [Beta]-glucuronidase activity using the substrate 4-methylumbellifery-[Beta]-D-glucuronide (MUG). When MUG is cleaved by the enzyme, a fluorescent product is produced that provides an intense blush-white fluorescence under longwave UV light. Approximately 92% of E coli isolates are MUG-positive. Almost all 0157:H7 and 0157:nonmotile isolates that produce Shiga-like toxins are MUG-negative.[1-3]
Toxin assay. Testing confirmed 0157:H7 isolates for Shiga-like toxins associated with pathogenicity is not routine. This is because there is an almost 100% correlation of toxin activity when the H7 antigen is present. Molecular methods such as pulsed-field gel electrophoresis (PFGE) and phage typing are necessary to confirm outbreaks of E coli 0157 since most strains are phenotypically similar. (See "A new tool for epidemiologists," p. 46, to learn how infection control officers and clinical laboratorians use PFGE to diagnose food-borne pathogens.)
NON-0157 ENTEROHEMORRHAGIC
E. COLI
Currently, E coli 0157:H7 is the most common of verotox-in-producing E coli serotypes. Various sources indicate it comprises from about 60% to more than 90% of all toxin-producing isolates and has been responsible for most outbreaks when food sources have been implicated.
Nevertheless, more than 60 other serotypes have been shown to be toxin-producers. Some of the more common ones appear to belong to somatic groups 0111, 026, 0104, 045, 0103, 0113, 0128, and 04. Serotypes 026:H11, 0111:NM (non-motile), and 0113:H21 appear to be isolated more frequently from patients with HUS.[2]
The absence of a biochemical marker for these other serotypes compared to E coli 0157:H7 (negative sorbitol reaction) and lack of any widespread use of antisera specific for their identification may lead to the underreporting of these organisms. Thus, E coli 0157:H7 statistics may be somewhat skewed since this is the one verotoxin-producer looked for on a wide-scale basis. A toxin assay in patients with bloody diarrhea and/or HUS with no isolation of 0157:H7 could be of significant value; they may be infected with a non-0157 toxin-producing E coli.
BEWARE OF PETS!
In September 1994, a five-month-old baby girl was hospitalized in New Jersey with acute illness, including fever, lethargy, vomiting, stiff neck, and a bulging from the soft spot on her head. Blood cultures and cerebral spinal fluid yielded Salmonella serotype Rubislaw. The baby was treated with intravenous ceftazidime for Salmonella sepsis and meningitis and discharged 10 days later.[4]
Salmonella serotype Rubislaw (Group F) is very rare and has been associated with reptiles, yet this baby's parents denied her having any contact with such a creature. Epidemiologic evidence indicated her babysitter did have an iguana (which she handled frequently) and that the child visited the house often. The pet was culture-positive for serotype Rubislaw.
A very high percentage of reptiles are asymptomatic carriers of Salmonella. Efforts to eliminate this pathogen from pet lizards have been largely unsuccessful. Since the mid-1980s, there has been a direct correlation between the increased popularity of iguanas and other reptiles as pets and an increased incidence of Salmonella infections due to reptile-associated serotypes.
The Centers for Disease Control and Prevention (CDC) recommends children, particularly those under the age of 5, limit their exposure to pet lizards. And, without a doubt, reptiles should never be brought into day care centers![4]
This pathogen probably remains the most important bacterial genus as far as gastrointestinal human disease and public health issues are concerned. It is routinely isolated from a large variety of domestic and wild animals, including chickens, birds, swine, cattle, rodents, reptiles, as well as pets such as dogs, cats, iguanas, tortoises, turtles, and terrapins. Transmission occurs by ingestion of the organisms in food derived from infected food-producing animals or contaminated by feces of an infected animal or person. Some outbreaks have been traced back to consumption of raw fruits and vegetables contaminated during preparation or by fertilizers and feeds. Among the most notorious sources: eggs and egg products, raw milk, contaminated water, meat, and poultry.
Eggs and poultry have led to various outbreaks. Salmonella serotype Enteritidis (SE), for instance, has been on the increase since the mid-1980s due to transovarian contamination of whole eggs. Estimates show one in 10,000 eggs contain SE. Outbreaks usually occur when Se-contaminated pooled eggs are undercooked or otherwise mishandled. Nursing homes are particularly vulnerable due to the overall debilitation of residents. SE continues to be the predominant serotype isolated in New Jersey.
Delineation of these outbreaks has allowed for proper control measures to be instituted, such as the following:
1. Substitute a pasteurized egg product when large quantities of eggs are needed and for institutional settings.
2. Discourage the use of raw eggs in foods such as hollandaise sauce, Caesar salad dressing, and homemade ice cream or eggnog.
3. Make the public aware that undercooked eggs pose a risk of SE infection.
According to both the CDC and the World Health Organization (WHO) Collaborating Center for Reference and Research on Salmonella in Paris, two species of Salmonella exist: Salmonella enterica and Salmonella bongori. These two species are further broken down into about 2,350 serotypes based on the combination of somatic (0) and flagellar (H) antigens present. Most of these serotypes belong to subspecies 1 and are given names.[5-6]
According to 1992 CDC statistics, 35,520 human source isolates were reported. Figure 2 lists the top 20 Salmonella serotypes with isolated numbers and percentages.
In some states in this country, regulations mandate the submission of cultures of Salmonella to the appropriate state laboratory for serotyping. Data on serotyping statistics is referred weekly to the CDC via the Public Health Laboratory Information System (PHLIS), which allows for rapid tracking of specific serotypes, particularly in cases of multistate outbreaks. (See "Tales from a state public health lab" for more information on Salmonella infection.)
TESTING FOR
SALMONELLA
SPECIES
Salmonella species are isolated from acutely ill patients on primary plating media, including MacConkey, EMB, Hektoen, XLD, Salmonella Shigella (SS), Brilliant Green, and Bismuth Sulfite; the latter two isolate Salmonella only. Enrichments include Tetrathionate (with or without Brilliant Green), Selenite, and Gramnegative broths (which must be used for testing convalescent patients or carriers.) Several different combined plating media can be used with these enrichments, for example, Hektoen, SS, as well as Brilliant Green.
Using Triple Sugar Iron Agar (TSI) and Lysine Iron Agar (LIA) in combination is an inexpensive screening technique. If reactions are typical for Salmonella, and agglutination with specific or polyvalent antisera is positive, there is a virtual 100% probability that the organism is a presumptive Salmonella.[7-8] multiple single-colony picks to TSI/LIA is recommended to increase isolation from convalescent patients; using multiple picks of similar colonies to any media, kit, or system is not.
MAJOR RISK TO PREGNANT WOMEN
In August 1985, an infectious outbreak was reported in Orange County, Calif., that linked a Mexican-style cheese to the onset of 142 cases of listeriosis. Almost two-thirds of the cases occurred in pregnant women or their offspring; about one-third of these cases ended in death. The cheese most likely was manufactured from a combination of raw and pasteurized milk The cheese plant that manufactured the incriminated cheese was found to be contaminated with Listeria monocytogenes, a Gram-positive rod that is found in soil, forage, water, mud, and animal feces.
These organisms attack the very young, the very old, pregnant patients and/or their fetuses, and the debilitated or immunocompromised. Its primary mode of transmission is through ingestion of contaminated foods, particularly nonreheated hot dogs, undercooked chicken, soft cheeses, and food purchased from delicatessens.
Typically the bacterial disease surfaces as fever, headache, and vomiting and progresses to meningoencephalitis, septicemia that may be accompanied by delirium, shock, and coma. The disease is particularly dangerous to pregnant women, whose infants, if infected, run the risk of being stillborn. The incubation period varies from 3-70 days-quite an extreme! Infected individuals may shed the organism in their stool for the duration o the illness.
EASY TO MISS,
DIFFICULT TO CONTROL
Listeria monocytogenes invades both animal and human tissues. Even though it grows well on commonly used clinical laboratory media, it can be confused as a streptococcus or diphtheroid and may be missed by a novice microbiologist. Isolation from clinical specimens typically steiile can be accomplished on blood agar plates incubated at 35 [degrees] C. Selective cultivation is suggested for contaminated specimens such as feces and vaginal secretions as well as for food products and other environmental specimens.
The University of Vermont (UVM) developed a commercially available medium and enrichment formulation (Oxoid, Unipath, Ogdensberg, N.Y.). The product is used in a two-step selective procedure that takes only three to four days versus older cold cultivations that take three months. Other enrichment formulations such as the one developed by the Netherlands Government Food Inspection Service (NGFIS), used in combination with UVM, can increase the serisitivity of the isolation procedure. The use of media such as Oxford agar and Polymyxin-acriflavine-lithium chloride-ceftazidime-esculin-mannitol (PALCAM) can simplify detection of Listeria, which have distinctive colonial morphology on these media. Immunological, PCR, and nucleicacid hybridization tests are available for food and environmental specimen screening.
Listeria monocytogenes can be identified by conventional biochemical tests, including gram stain, hemolysis, CAMP test, and motility; DNA probe (Gen Probe, San Diego, Calif.) and for environmental and food specimen sources API-Listeria (biomerieux Vitek, Hazelwood, Mo.). Identification is especially important from the latter since all Listeria species may be isolated from these. Outbreak-related specimens may be serotyped, but more discriminatory subtyping such as phage typing, multilocus enzyme electrophoresis, and DNA fingerprinting must be used since most human disease is caused by serotypes 1/2a, 1/2b, and 4b.[9]
Listeria is a very difficult organism to control in food since it:
* is ubiquitous in nature
* is relatively resistant to heat
* can tolerate high-sait concentrations
* survives freezing
* thrives in all processed foods
* is a psychotropic pathogen, that is, able to grow rapidly in a refrigerated environment.
The organism is the masked bandit of bacterial pathogens in that it can grow to truly high concentrations in refrigerated conditions without obvious signs of contamination (i.e., foul odor, altered appearance of food).
The Food and Drug Administration and the U.S. Department of Agriculture have imposed a zero tolerance for the presence of Listeria in all processed dairy products and ready-to-eat foods. This aggressive program of prevention, education, and regulation may have tumed the tide on the rising rate of human listeriosis in the 1980s to a noticeable decline in the past few years. Nevertheless, microbiologists must be aware of this pathogen and ready to educate others to its significance.
WHAL CAUSED THAT ULCER?
Once upon a time, modern medicine believed peptic ulcers were caused by stress, diet, smoking, and Type A personality. Wrong! A spiral-shaped, Gram-negative bacterium with four to six flagella found in the mucous layer of the stomach or duodenum is the culprit. Dr. Barry J. Marshall of the Department of Medicine at the University of Virginia Health Sciences Center in Charlottesville discovered the connection between Helicobacter pylori and peptic ulcer, however, many in the medical community were skeptical and it was difficult to prove the theory without the right animal model. Consequently, in a dramatic demonstration, Marshall infected himself with a pure H. pylori culture and subsequently experienced severe acute gastritis with hypochlorhydria. This condition was linked to H. pylori infection, which was then confirmed by culture.
This incident opened the way to treating peptic ulcer with bismuth compounds and a combination of antibiotics such as metronidazole, ampicillin, and tetracycline. The goal is to totally eradicate this organism, which should coincide with complete recuperation.
H. pylori has emerged as the recognized pathogen for most individuals suffering from peptic ulcer disease. In underdeveloped countries, more than 75% of adults are infected with this disease. In the U.S. as well as other developed countries, as few as 20% of adults are infected. Those immigrating into this country from developing regions are at much greater risk of developing peptic ulcer; however, it is extremely exciting to think that we have developed an effective treatment for a new pathogen that has the potential to control and reduce peptic ulcer disease quite substantially.
The exact mode of H. pylori transmission is still unknown, and yet it is assumed that the infection is acquired during childhood, carried through life, and related to exposure to contaminated food and water. Persons born postseem 1950 to have a lower prevalence of infection than those who are born before this year. Most people colonized with H. pylori are asymptomatic, and possibly only about 1% of infected adults develop peptic ulcers. The answer to who will develop peptic ulcer appears to be linked to the production of a soluble cytotoxin by the organism, which may induce an inflammatory reaction, thereby inducing ulcer.
Though many details remain unclear at this time, participants at the National Institutes of Health Consensus Conference held in February 1995 predicted that, since the cure rate of peptic ulcer is high and treatment relatively simple, up to 90% of H. pyloybrelated peptic ulcer diseases in the U.S. may be eliminated.
TESTING FOR H. PYLORI
H. pyloti, an etiologic agent of chronic central gastritis, is a fastidious organism. Even though it is unable to grow in regular blood culture systems, it can be recovered in specialized media developed to isolate Brucella. These include Brucella broth with [CO.sub.2] and SPS, Biphasic Brain-Heart Infusion agar or broth, and SP broth. Incubation should be up to 14 days for the highest recovery; however, the majority of organisms will be isolated after seven days.
Clinical specimens including biopsy material, dental plaque, and saliva should be plated on both selective and nonselective media. Chocolate agar or Brucelia agar supplemented with 5%-7% horse or rabbit blood and one of a variety of specialized selective media including Dent's medium (Oxoid), Pylori agar (bioMerieux, Marcy I'Etoile, France) and modified Skirrow's media with added amphotericin B and cefsulodin replacing polymyxin are used.[10] Incubation is up to seven days on solid media; most isolates are recovered after four.
Identification methods include convendonal biochemical tests. Presumptive identification can be based on typical helical, curved, or straight rods, 0.3-1.0 um wide to 1.5-5.0 um long and positive catalase, oxidase, and rapid urease reactions.[11] The difficulty of invasive biopsy culture has led to the development of commercial, noninvasive diagnostic tests for H. pylori antibodies. These assays test for the H. pylodspecific serum immunoglobulin (IgG).
A study of five commercial tests found patient population affects the sensitivity and specificity of these tests. Researchers much greater correlation between infection and seropositivity in a group under the age of 45. Laboratories using tests must base the selection of a test kit on the evalation of the diagnostic accuracy in a selected population and on cost and numbers being tested.[12]
SOUND THE
ALARM
Intestinal infections cost the U.S. an estimated $23 billion a year in direct medical costs and lost productivity. Food-borne diseases probably account for a large slice of that pie, and the majority of cases may remain unreported, thereby putting additional lives at risk.
Many regional and national groups currently are working with the CDC to institute a national standardized system for the electronic reporting of communicable diseases and other critical health-related information to eventually form a network for global surveillance. This should revolutionize strategies to prevent the spread of disease and save countless lives due to the early warning.
Part of what keeps our standard of living the envy of the world is our high criterion of public health. Every clinical laboratorian must take a leadership role for sounding the alarm and reporting communicable diseases promptly. Let us never forget our role in maintaining and improving the public health of our community as well as others around the globe. (See Figure 3 for tips on avoiding the spread of food-bome disease in the kitchen.)
References
[1.] Centers for Disease Control and Prevention. National Center for Infectious Diseases. E coli 0157:H7: What the Clinical Microbiologist Should Know. 1994.
[2.] Griffin P, Tauxe RV. The epidemiology of infections caused by Escherichia coli 0157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiologic Reviews. 1199; 13:60-98
[3.] Karmali MA. Infection by verocytotoyin-producing Escherichia coli. Clin Microbiol Review. 1989; 2:15-38.
[4.] Centers for Disease Control and Prevention. Reptile-associated Salmonellosis -- Selected States, 1994-1995. MMWR. 44:347-350.
[5.] McWhorter-Murlin AC, Hickman-Brenner FW. Identification and Serotyping of Salmonella, and an Update of the Kauffinann-White Scheme Atlanta, Ga: Centers for Disease Control and Prevention; 1994.
[6.] Popoff MY, LeMinor L. Antigenic Formulas of the Salmonella Serovars. Institute Pasteur, Paris: World Health Organization Collaborating Center for Reference and Research on Salmonella; 1992.
[7.] Balows A, ed. Manual of Clinical Microbiology. 5th ed. Washington, DC: American Society for Microbiology; 1991; Chapter 36.
[8.] Farmer JJ, Wells JG, Griffin PM, Wadismuth IK Diagnostic Procedures for Bactirial Infections. 7th ed. Washington, DC: American Public Health Association Inc.; 1987:233-296, chap 14.
[9.] Food-bome microorganisms and their toxins. In: Pierson MD, Stem NJ, eds. Developing Methodology. New York, NY: Marcel Dekker, Inc.; 1986.
[10.] Kehler EG, Midlaff BR, Westblom TU. Evaluation of three commercially available blood culture systerns for the cultivation of Helicobacter pylori. J Clin Microbiol. 1994; 32:1,597-1,598.
[11.] Manual of Clinical Microbiology. 6th ed. In: Murray P, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, eds. Washington, DC: ASM Press; 1995.
[12.] Schembri MA, Lin SK, Lambert JR. Comparison of commercial diagnostic tests for Helicobacter pylori antibodies. J Clin Microbiol. 1993; 31:2,621-2,624.
Tales from a state public health lab
Infectious organisms do not respect state or national borders; they enter a once-infection-free area as easily as stepping off an airplane. One such outbreak occurring last year involved two rare Salmonella serotypes: Hartford (Group [C.sub.1]) and Gaminara (Group I). (Both serotypes comprise less than 0.3% of the total isolates of any given year.) New Jersey sounded the warning bell when a cluster of Salmonella sertype Hartford cases were reported to the CDC in June 1995. All were received at our lab during the first and second weeks of June from seven different referral agencies, representing both hospital and private labs. One of the last cultures received in mid-June, although referred for Salmonella-[C.sub.1]-serotyping, was a mixed culture of both Hartford and Gaminara. It was pure serendipity that when this culture was streaked to MacConkey agar for purity confirmation, the first picks made were Group I positive, not group [C.sub.1] and I.
As the outbreak progressed, epidemiological evidence from Mew Jersey and Florida isolates traced the infections back to travel to a popular family theme park in Central Florida as well as to the consumption of an upasteurized orange juice there. Eventually some 60 cases of Salmonella serotype Hartford from 21 different states met the case definition as delineated by epidemiologists.
The orange juice from a specific processor was cultured for Salmonella, and serotype Gaminara was isolated from both opened and unopened containers. While serotype Hartford was not isolated from the juice, epidemiologic data and the presence of serotype Gaminara in both the juice and patients implicated an ongoing Salmonella contamination from the processor that supplied the juice to the park during the time when patients vacationed there. Cooperation among various health-related agencies was necessary to describe the extent of the outbreak, track its source, and ultimately institute controls for containment.(*)
Another serotype of Salmonella was discovered in two human cases in March 1995. The organism was first isolated from the appendix of an adult female and then from a spinal fluid of a five-month-old infant, unfortunately resulting in her death. The antigenic formula of the isolate is 028:b:l,w (serogroup M).
Since this organism is a member of subspecies 1, our facility was given the honor of naming it. Salmonella serotypes are named after the geographic area where initial strains are first isolated. Since one of these patients was from Tinton Falls and the other from Freehold (both in New Jersey), the names were combined to create Salmonella serotype Freefalls, which was subsequently accepted by the WHO.
(*) source: Cook KA. Memorandum Outbreak of Salmonella Hartford Infections Among Travelers to Orlando, Florida, May 1995. Atlanta, Ga: Centers for Disease Control and Prevention; 1995.
Tips on avoiding the spread of
foodborne disease in the kitchen
* Thoroughly cook meat, chicken, and fish. The center of such meat should not be pink. Cook hamburgers until their juices run clear.
* Avoid recipes that use raw or undercooked eggs. For instance, homemade ice cream, cookie batter, Caesar salad dressing, and hollandaise sauce may harbor Salmonella.
* Wash your kitchen chopping board and utensils, as well as your hands, with hot, soapy water every time you touch raw meat or poultry.
* Wrap uncooked meat and poultry before storing in the refrigerator.
* Thaw frozen meat in the refregirator.
* Thoroughly wash all fruits and vegetables. They may be contaminated with harmful organisms found in the soil or fertilizers.
* Refrain from cleaning reptile cages in kitchens and other food preparation areas.
COPYRIGHT 1996 Nelson Publishing
COPYRIGHT 2004 Gale Group